In the realm of biology, observation can foster comprehension, and scientists in Professor Edward Boyden’s laboratory at the McGovern Institute for Brain Research are dedicated to elucidating life with greater clarity. Utilizing two innovative methodologies, they are enhancing the functionalities of expansion microscopy — a cutting-edge imaging process introduced by the team in 2015 — enabling researchers globally to perceive more when examining cells and tissues through a light microscope.
“Our aspiration is to observe everything; hence, we are continuously seeking enhancements,” remarks Boyden, the Y. Eva Tan Professor in Neurotechnology at MIT. “Capturing a comprehensive image of all life, down to its essential components, is essentially our aim.” Boyden also holds a position as a Howard Hughes Medical Institute investigator and is a participant in the Yang Tan Collective at MIT.
With novel techniques for staining samples and processing images, practitioners of expansion microscopy can now discern vivid delineations of cell shapes in their images and accurately identify the locations of a variety of proteins within a single tissue specimen with a resolution that significantly surpasses conventional light microscopy. These improvements, both documented in open-access format in the journal Nature Communications, enable fresh methods for tracing the slender extensions of neurons and visualizing spatial relationships among molecules that influence health and disease.
Expansion microscopy employs a hydrogel that absorbs water to physically enlarge biological tissues. Once a tissue sample has been saturated with the hydrogel, it is hydrated. The hydrogel expands as it takes in water, maintaining the relative positions of molecules in the tissue while gradually pulling them apart. Consequently, densely packed cellular components appear separate and distinct when the expanded tissue is examined under a light microscope. This technique, which can be executed using standard lab equipment, has rendered super-resolution imaging attainable for most research groups.
Since the initial development of expansion microscopy, Boyden and his team have persistently refined the method — enhancing its resolution, streamlining the procedure, creating new features, and integrating it with other tools.
Visualizing cellular membranes
Among the team’s most recent advancements is a technique known as ultrastructural membrane expansion microscopy (umExM), discussed in the February 12 issue of Nature Communications. This enables biologists to utilize expansion microscopy to visualize the delicate membranes that delineate cells and encase organelles within them. These membranes, primarily composed of molecules termed lipids, have been notoriously challenging to densely label in intact tissues for imaging via light microscopy. Now, scientists can apply umExM to examine cellular ultrastructure and organization in tissues.
Tay Shin SM ’20, PhD ’23, a former graduate student in Boyden’s lab and a J. Douglas Tan Fellow at the Tan-Yang Center for Autism Research at MIT, spearheaded the development of umExM. “Initially, our objective was quite straightforward: Let’s label membranes in intact tissue, similar to how an electron microscope uses osmium tetroxide to highlight membranes for tissue visualization,” he states. “It turns out achieving this is exceedingly difficult.”
The team initially needed to create a label that would render the membranes in tissue samples visible under light microscopy. “We practically had to start from the beginning,” Shin explains. “We had to critically analyze the fundamental properties of the probe intended for labeling the plasma membrane and then consider how to integrate those into expansion microscopy.” This involved engineering a molecule that would attach to the lipids composing the membrane and connect it to both the hydrogel used for expanding the tissue sample and a fluorescent molecule for visibility.
After improving the expansion microscopy protocol for membrane visualization and thoroughly testing various probes, Shin achieved success one late night in the lab. He placed an expanded tissue sample on a microscope and observed sharp outlines of cells.
The heightened resolution provided by expansion allowed Boyden’s team to identify even the minuscule dendrites extending from neurons and clearly visualize the elongated projections of their slender axons. Such precision could assist researchers in tracing individual neurons’ trajectories within the densely interconnected networks of the brain, the researchers assert.
Boyden describes tracking these neural pathways as “a top priority of our era in brain science.” Traditionally, such tracing has relied heavily on electron microscopy, which necessitates specialized expertise and costly equipment. According to Shin, because expansion microscopy utilizes a standard light microscope, it is significantly more accessible to laboratories worldwide.
Shin and Boyden note that users of expansion microscopy can gain even deeper insights into their samples when they combine the new capability to illuminate lipid membranes with fluorescent labels indicating the locations of specific proteins. “That’s crucial, as proteins perform many functions within the cell, but it’s important to know their locations relative to the cell’s structure,” states Boyden.
One sample, multiple proteins
To facilitate this, researchers no longer need to limit their selection to just a few proteins when employing expansion microscopy. Through a new method termed multiplexed expansion revealing (multiExR), users can now label and visualize over 20 distinct proteins within a single sample. Biologists can use this method to observe groups of proteins, comprehend their organizational structure relative to one another, and formulate new hypotheses regarding their interactions.
A vital aspect of this new method, reported on November 9, 2024, in Nature Communications, is the capacity to repeatedly attach fluorescently labeled antibodies to specific proteins in an expanded tissue sample, image them, then remove these antibodies and apply a new set to unveil another array of proteins. Postdoc Jinyoung Kang refined each phase of this procedure, ensuring tissue samples remained intact and the labeled proteins yielded bright signals during each imaging session.
Following the acquisition of numerous images from a single sample, Boyden’s team encountered an additional challenge: ensuring that these images were perfectly aligned for overlaying, thereby generating a final image that illustrated the exact positions of all the labeled and visualized proteins, one by one.
Expansion microscopy enables biologists to visualize some of the minutest features of cells — but to locate the same features consistently across multiple imaging rounds, Boyden’s team first needed to focus on a larger structure. “These areas of view are incredibly tiny, and you’re attempting to find this exceptionally small area of view in a gel that has significantly expanded,” clarifies Margaret Schroeder, a graduate student in Boyden’s lab who, alongside Kang, led the development of multiExR.
To consistently navigate to the correct location, the team opted to label the blood vessels traversing each tissue sample and utilize these as a reference point. For precise alignment, certain subtle details had to be consistently visible in every image; for this purpose, the team labeled several structural proteins. With these reference markers and custom imaging processing software, the team successfully integrated all of their images of a sample into one, revealing the spatial arrangement of proteins that had been visualized independently.
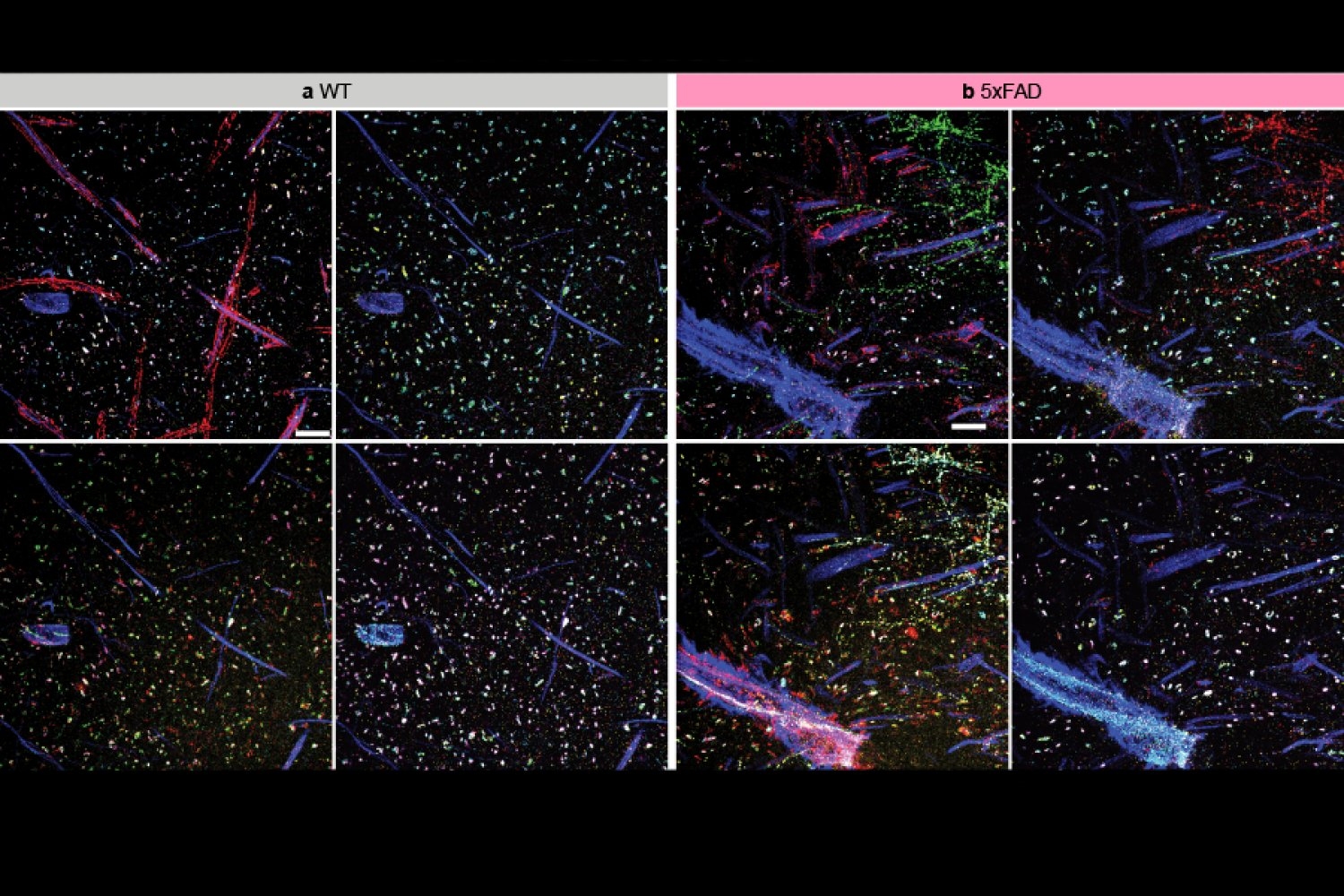
The team employed multiExR to investigate amyloid plaques — the abnormal protein aggregates that notoriously form in the brains of individuals with Alzheimer’s disease. “We could delve into those amyloid plaques and inquire, what’s contained within them? And because we can stain for numerous proteins, we could conduct a high-throughput analysis,” Boyden states. The team selected 23 different proteins to examine in their images. This approach unveiled some unexpected findings, such as the detection of specific neurotransmitter receptors (AMPARs). “Here’s one of the most renowned receptors in neuroscience, and there it is, tucked away in one of the most recognized molecular indicators of pathology in neuroscience,” remarks Boyden. The role, if any, that these receptors play in Alzheimer’s disease remains uncertain — but this discovery demonstrates how the capability to examine more within cells can reveal surprising aspects of biology and provoke new lines of inquiry for research.
Funding for this research was provided by MIT, Lisa Yang and Y. Eva Tan, John Doerr, the Open Philanthropy Project, the Howard Hughes Medical Institute, the U.S. Army, Cancer Research U.K., the New York Stem Cell Foundation, the U.S. National Institutes of Health, Lore McGovern, Good Ventures, Schmidt Futures, Samsung, MathWorks, the Collamore-Rogers Fellowship, the U.S. National Science Foundation, Alana Foundation USA, the Halis Family Foundation, Lester A. Gimpelson, Donald and Glenda Mattes, David B. Emmes, Thomas A. Stocky, Avni U. Shah, Kathleen Octavio, Good Ventures/Open Philanthropy, and the European Union’s Horizon 2020 program.