A comprehensive exploration of natural variety has resulted in researchers at MIT’s McGovern Institute for Brain Research and the Broad Institute of MIT and Harvard discovering ancient systems that hold promise for enhancing the genome editing toolkit.
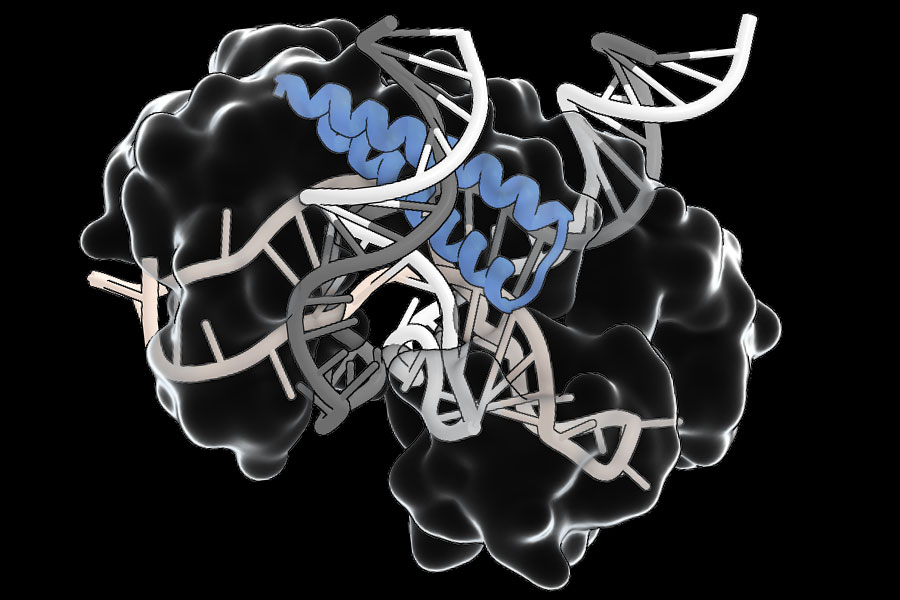
These systems, referred to as TIGR (Tandem Interspaced Guide RNA) systems by the scientists, utilize RNA to direct them to specific locations on DNA. TIGR systems can be reconfigured to focus on any DNA sequence of interest, and they possess unique functional components that can operate on the targeted DNA. Besides its modular nature, TIGR is considerably smaller in comparison to other RNA-guided systems, like CRISPR, which is a significant advantage for its therapeutic delivery.
These results are discussed online Feb. 27 in the journal Science.
“This is an exceptionally adaptable RNA-guided system with numerous diverse functions,” states Feng Zhang, the James and Patricia Poitras Professor of Neuroscience at MIT, who spearheaded the study. The TIGR-associated (Tas) proteins that Zhang’s group identified include a common RNA-binding element that interacts with an RNA guide pinpointing a specific location in the genome. Some of these proteins cleave the DNA at that position, utilizing a nearby DNA-cutting segment of the protein. This modularity could streamline tool development, enabling researchers to incorporate advantageous new capabilities into natural Tas proteins.
“Nature is quite remarkable,” remarks Zhang, who is also an investigator at the McGovern Institute and the Howard Hughes Medical Institute, a core member of the Broad Institute, a professor of brain and cognitive sciences and biological engineering at MIT, and co-director of the K. Lisa Yang and Hock E. Tan Center for Molecular Therapeutics at MIT. “It offers an immense diversity, and we have been probing that natural variety to discover new biological mechanisms and harnessing them for diverse applications to manipulate biological processes,” he adds. Prior to this, Zhang’s group modified bacterial CRISPR systems into gene editing instruments that have revolutionized contemporary biology. His group has also uncovered a plethora of programmable proteins from CRISPR systems and beyond.
In their latest study aimed at discovering novel programmable systems, the team initially concentrated on a structural characteristic of the CRISPR-Cas9 protein that connects to the enzyme’s RNA guide. This is a crucial feature that has rendered Cas9 an exceptionally effective tool: “Being RNA-guided simplifies the reprogramming process, as we understand how RNA associates with other DNA or other RNA,” Zhang clarifies. His team analyzed hundreds of millions of biological proteins with known or predicted structures, seeking out any that exhibited a similar domain. To identify more distantly related proteins, they employed an iterative approach: from Cas9, they pinpointed a protein called IS110, previously established by others to bind RNA. They then focused on the structural properties of IS110 that permit RNA binding and reiterated their search.
At this stage, the search uncovered such a vast number of distantly related proteins that the team resorted to artificial intelligence to interpret the list. “When undertaking iterative, deep mining, the resulting findings can be so varied that they become challenging to analyze using standard phylogenetic techniques, which depend on conserved sequences,” explains Guilhem Faure, a computational biologist in Zhang’s laboratory. With a large protein language model, the team managed to categorize the proteins they identified into clusters based on their likely evolutionary connections. One group stood out from the others, with its members being particularly captivating because they were encoded by genes displaying regularly spaced repetitive sequences analogous to a vital component of CRISPR systems. These were the TIGR-Tas systems.
Zhang’s team identified over 20,000 distinct Tas proteins, primarily found in viruses that infect bacteria. The sequences within each gene’s repetitive domain — its TIGR arrays — encode an RNA guide that engages with the RNA-binding section of the protein. In some instances, the RNA-binding region is next to a DNA-cleaving section of the protein. Others seem to associate with different proteins, indicating they may assist in directing those proteins to DNA targets.
Zhang and his team experimented with numerous Tas proteins, proving that several can be programmed to create targeted cuts in DNA within human cells. As they contemplate developing TIGR-Tas systems into programmable tools, the researchers are optimistic about features that may render these tools particularly adaptable and precise.
They observe that CRISPR systems can only target segments of DNA flanked by short motifs known as PAMs (protospacer adjacent motifs). In contrast, TIGR Tas proteins possess no such restriction. “This implies that theoretically, any location in the genome should be targetable,” states scientific advisor Rhiannon Macrae. Additionally, the team’s experiments reveal that TIGR systems feature what Faure describes as a “dual-guide system,” interacting with both strands of the DNA double helix to accurately hone in on their target sequences, ensuring that they only operate where guided by their RNA guide. Furthermore, Tas proteins are compact — an average of one-fourth the size of Cas9 — facilitating their delivery, which may help surmount a significant barrier to the therapeutic application of gene-editing tools.
Enthusiastic about their discovery, Zhang’s group is now investigating the natural function of TIGR systems in viruses, as well as their applicability to research or therapeutics. They have deciphered the molecular structure of one of the Tas proteins shown to function in human cells and intend to use that information to enhance its efficiency. Moreover, they note connections between TIGR-Tas systems and specific RNA-processing proteins in human cells. “I believe there is more to explore regarding what some of those connections may be, and it might help us gain a better understanding of how these systems are utilized in humans,” says Zhang.
This research was supported by the Helen Hay Whitney Foundation, Howard Hughes Medical Institute, K. Lisa Yang and Hock E. Tan Center for Molecular Therapeutics, Broad Institute Programmable Therapeutics Gift Donors, Pershing Square Foundation, William Ackman, Neri Oxman, the Phillips family, J. and P. Poitras, and the BT Charitable Foundation.