When Xiao Wang submitted applications for academic positions, numerous institutions she interviewed with perceived her research proposal — focused on examining the life cycle of RNA in cells and its impact on normal development and ailments — as overly expansive.
Nonetheless, this perception was not shared during her interview at MIT, where her prospective colleagues welcomed her concepts and urged her to be even more daring.
“What I am currently engaged in is even more extensive, even more audacious than my original proposal,” states Wang, who has dual appointments in the Department of Chemistry and the Broad Institute of MIT and Harvard. “I received tremendous support from all my peers in my department and at Broad, enabling me to acquire the resources necessary for pursuing my research goals. This also exemplifies the courage of the students here. There exists a truly innovative culture and atmosphere, so students are not deterred by projects that might seem unusual or unattainable.”
Wang’s investigation into RNA unites scholars from chemistry, biology, computer science, neuroscience, and other disciplines. Within her laboratory, research concentrates on creating tools that identify where various types of messenger RNA are translated into proteins within a specific cell — data that can provide insights into how cells determine their fate and the abnormalities that occur in diseases, particularly in the brain.
“The collaborative role between MIT Chemistry and the Broad Institute was particularly appealing to me as I was trained as a chemist and wished to teach and attract students from chemistry. Simultaneously, I desired exposure to biomedical subjects and to collaborate with individuals outside of chemistry. I can partner with biologists, physicians, and computational scientists who analyze these extensive datasets,” she notes.
Imaging RNA
Wang commenced her tenure at MIT in 2019, just prior to the onset of the Covid-19 pandemic. Until that moment, she was hardly acquainted with anyone in the Boston region, yet she received a warm reception.
“I wasn’t educated at MIT, and I had never lived in Boston previously. Initially, my social circles were quite limited, consisting solely of my colleagues and students, yet remarkably, even during the pandemic, I never felt socially isolated. I felt integrally connected even though it was a tight-knit circle,” she explains.
Having grown up in China, Wang developed an interest in science during middle school, when she was selected to participate in China’s National Olympiad in mathematics and chemistry. This experience allowed her to engage with college-level material, culminating in her winning a gold medal in the nationwide chemistry competition.
“That exposure was enough to initially draw me into mathematics and later toward chemistry. That’s how I became interested in pursuing a more science-focused major and career,” Wang reflects.
At Peking University, she specialized in chemistry and molecular engineering. While there, she collaborated with Professor Jian Pei, who provided her with the opportunity to independently manage her own research endeavor.
“I have a genuine passion for research because every day you formulate a hypothesis, design an experiment, and bring it to fruition. It resembles playing a video game: You enjoy a daily feedback loop, sometimes receiving rewards, sometimes not. It feels significantly more engaging than attending classes, which led me to decide to apply for graduate school,” she shares.
As a graduate student at the University of Chicago, she became captivated by RNA while completing a rotation in Chuan He’s laboratory, a chemistry professor. He investigated chemical modifications that influence the function of messenger RNA — the molecules that convey protein-synthesis instructions from DNA to ribosomes, the sites of protein assembly.
Wang ultimately joined He’s lab, where she investigated a prevalent mRNA modification, m6A, which affects the efficiency of mRNA translation into protein and the rate of its degradation within the cell. Furthermore, she began examining how mRNA modifications influence embryonic development. For these studies, she utilized zebrafish, which possess transparent embryos developing from fertilized eggs into free-swimming larvae within a mere two days. This piqued her interest in creating techniques that could show where different RNA types were expressed via imaging the organism as a whole.
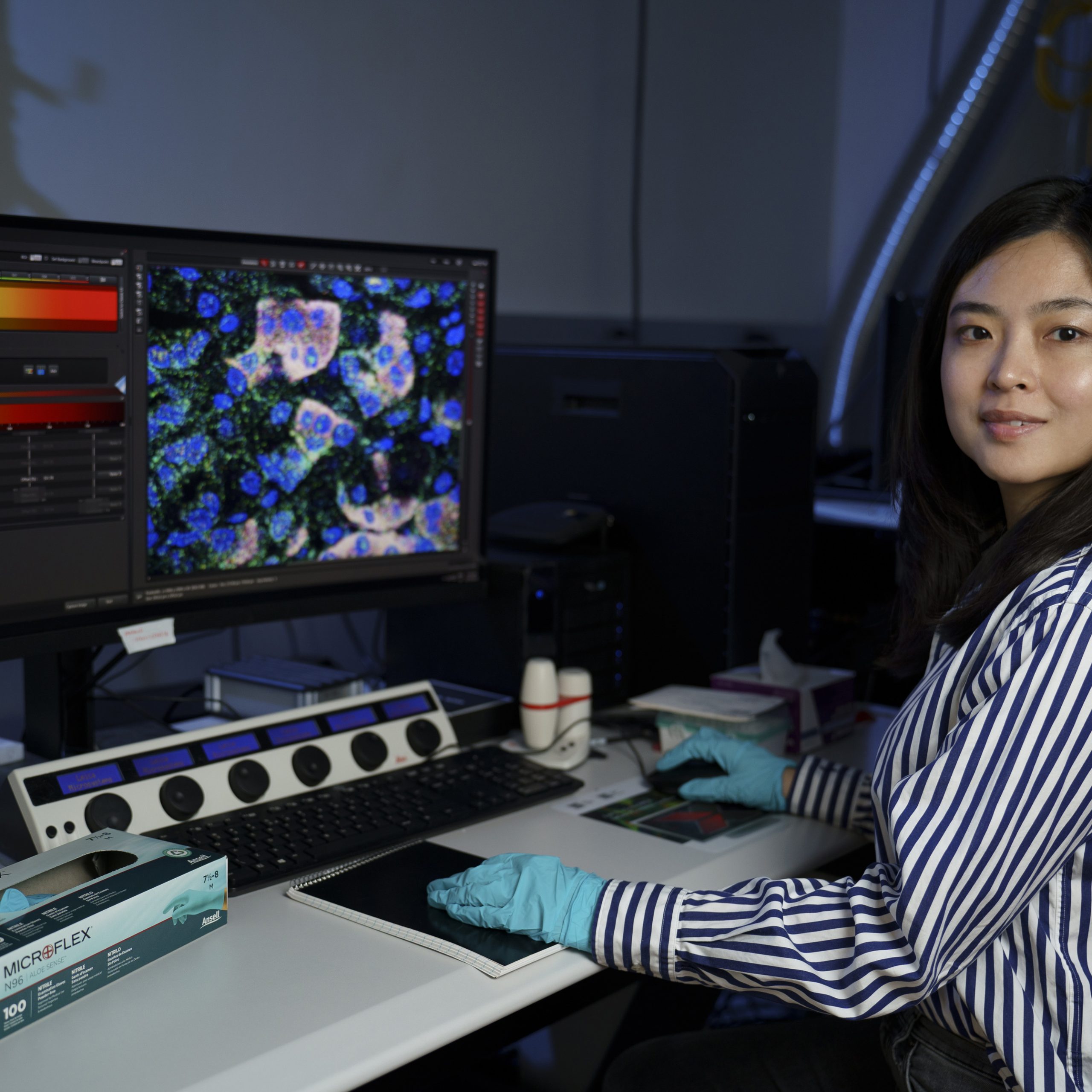
She quickly recognized that such an approach could also benefit brain research. During her postdoc at Stanford University, she commenced developing RNA imaging techniques in collaboration with Professor Karl Deisseroth. Although there are established methods for identifying mRNA molecules expressed in individual cells, these do not provide precise locations within the cells of different mRNA types. She initiated the development of a method called STARmap, designed to achieve this form of “spatial transcriptomics.”
Using this methodology, researchers initially employ formaldehyde to crosslink all mRNA molecules in situ. Next, the tissue is treated with fluorescent DNA probes that are complementary to the target mRNA sequences. These probes can subsequently be imaged and sequenced, revealing the locations of each mRNA sequence within a cell. This facilitates the visualization of mRNA molecules encoding thousands of different genes within single cells.
“I was harnessing my knowledge in RNA chemistry to create this RNA-focused brain mapping technology, enabling the use of RNA expression profiles to define types of brain cells and to visualize their spatial architecture,” Wang explains.
Tracking the RNA life cycle
Wang’s lab members are now engaged in enhancing the capabilities of the STARmap technique to facilitate analyses of brain function and neural connectivity. They are also creating tools to map the entire life cycle of mRNA molecules, from synthesis to translation to degradation, and to monitor how these molecules are transported within cells throughout their lifespan.
One of these tools, known as RIBOmap, identifies the locations of mRNA molecules during their translation at ribosomes. Another tool allows researchers to assess the speed at which mRNA degrades following transcription.
“We are striving to develop a toolkit that will permit us to visualize each stage of the RNA life cycle within cells and tissues,” Wang notes. “These represent newer generations of tool development focused on crucial RNA biological inquiries.”
One central question involves how different cell types uniquely regulate their RNA life cycles and the subsequent effects on their differentiation. Variations in RNA regulation might also contribute to diseases such as Alzheimer’s. In a 2023 study, Wang and MIT Professor Morgan Sheng utilized a version of STARmap to unveil how microglia cells become increasingly inflammatory as amyloid-beta plaques form within the brain. Wang’s lab is also conducting research into how disparities in mRNA translation could impact schizophrenia and other neurological conditions.
“We believe that there is a wealth of fascinating biology awaiting discovery because the formation of neural circuits occurs through synapses, and synapse formation, along with learning and memory, is closely tied to localized RNA translation, which involves numerous steps, including RNA transport and recycling,” she states.
In addition to delving into those biological inquiries, Wang is also exploring methods to enhance the efficacy of mRNA therapeutics and vaccines by altering their chemical modifications or their topological configuration.
“Our objective is to construct a toolbox and RNA synthesis strategy that allows us to precisely modify the chemical attributes of every RNA molecule,” Wang explains. “We aim to elucidate how these modifications influence the speed at which mRNA can generate protein and in which cell types they may be utilized to enhance protein production efficiency.”