“`html
For individuals afflicted with inflammatory bowel disease, antibiotics can be a double-edged sword. The broad-spectrum medications frequently administered for gastrointestinal flare-ups can eradicate beneficial microbes along with harmful ones, occasionally intensifying symptoms over time. When combating gut inflammation, you don’t always want to approach with a hammer rather than a scalpel.
Investigators at MIT’s Computer Science and Artificial Intelligence Laboratory (CSAIL) and McMaster University have discovered a novel compound that adopts a more focused strategy. The molecule, named enterololin, inhibits a specific group of bacteria associated with Crohn’s disease flare-ups while keeping the majority of the microbiome largely unharmed. Utilizing a generative AI model, the team charted how the compound operates, a procedure that typically takes years but was expedited here to merely months.
“This finding addresses a fundamental obstacle in antibiotic advancement,” notes Jon Stokes, senior author of a recent publication on the study, assistant professor of biochemistry and biomedical sciences at McMaster, and research affiliate at MIT’s Abdul Latif Jameel Clinic for Machine Learning in Health. “The challenge isn’t uncovering molecules that eliminate bacteria in vitro — we’ve been adept at that for a long time. A significant barrier lies in understanding what those molecules actually accomplish within bacteria. Without that nuanced comprehension, you cannot convert these early-stage antibiotics into safe and effective treatments for patients.”
Enterololin represents a move towards precision antibiotics: therapies aimed at eliminating only the problematic bacteria. In murine models of Crohn’s-like inflammation, the medication specifically targeted Escherichia coli, a gut-residing bacterium that can exacerbate flare-ups, while leaving most other microbial inhabitants untouched. Mice administered enterololin exhibited more rapid recovery and maintained a healthier microbiome than those treated with vancomycin, a widely used antibiotic.
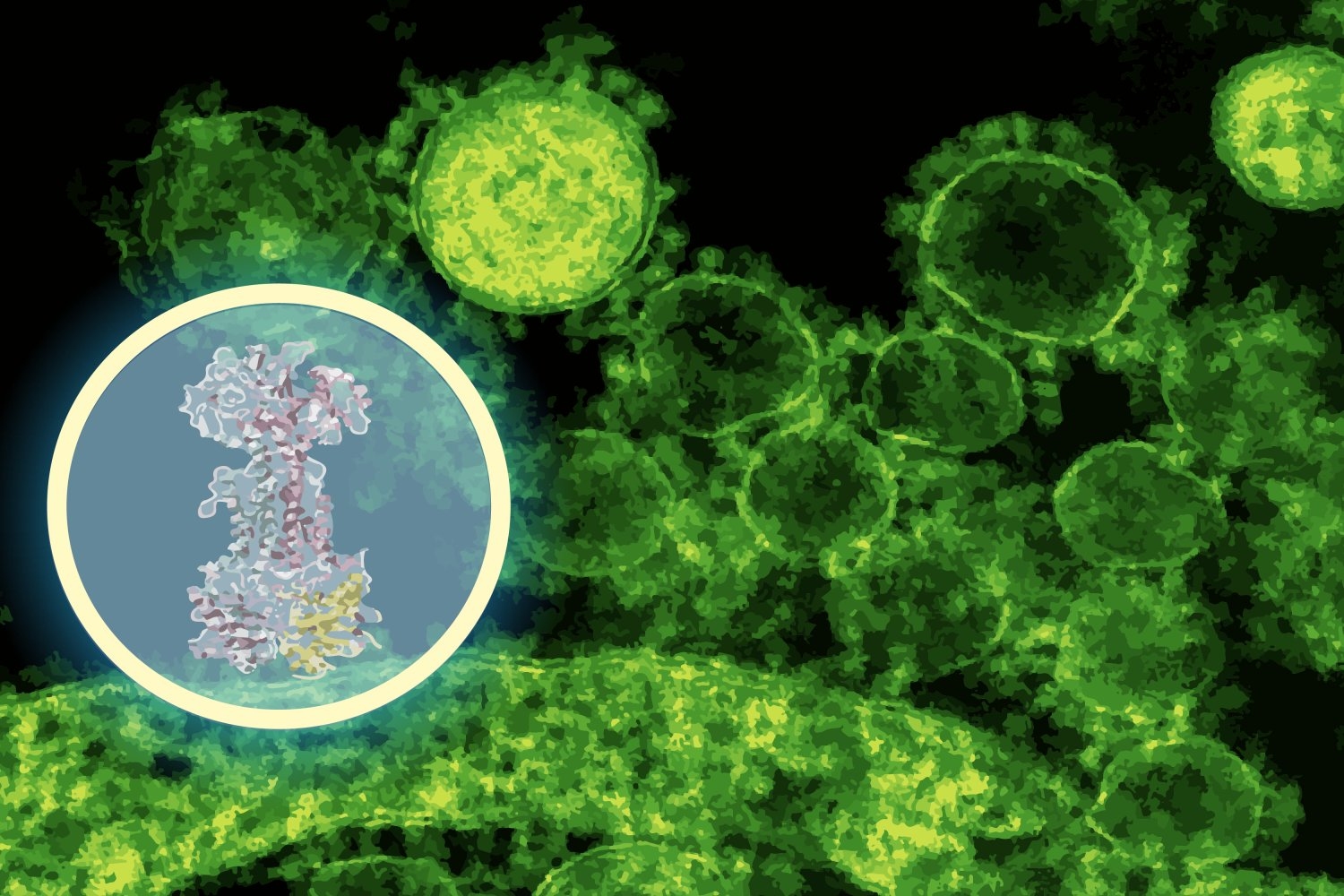
Identifying a drug’s action mechanism, the molecular target with which it interacts inside bacterial cells, generally necessitates years of meticulous experimentation. Stokes’ lab discovered enterololin via a high-throughput screening technique, but pinpointing its target would have been the limiting factor. Here, the team utilized DiffDock, a generative AI model developed at CSAIL by MIT PhD student Gabriele Corso and MIT Professor Regina Barzilay.
DiffDock was developed to foresee how small molecules fit into the binding sites of proteins, a notoriously challenging problem in structural biology. Conventional docking algorithms navigate through potential orientations using scoring rules, often yielding noisy outcomes. DiffDock, on the other hand, reframes docking as a probabilistic reasoning dilemma: a diffusion model iteratively refines predictions until it settles on the most probable binding configuration.
“In just a few minutes, the model predicted that enterololin binds to a protein complex known as LolCDE, essential for transporting lipoproteins in specific bacteria,” states Barzilay, who also co-heads the Jameel Clinic. “That was a substantial lead — one that could provide guidance for experiments, rather than replace them.”
Stokes’ team then validated that prediction. Using DiffDock predictions as an experimental guide, they initially evolved enterololin-resistant mutants of E. coli in the laboratory, which indicated that modifications in the mutant’s DNA were linked to lolCDE, exactly where DiffDock had anticipated enterololin would bind. They also conducted RNA sequencing to observe which bacterial genes activated or deactivated upon exposure to the drug, as well as employed CRISPR to selectively suppress expression of the predicted target. These laboratory trials consistently revealed disturbances in pathways related to lipoprotein transport, exactly as DiffDock had forecasted.
“When you witness the computational model and the experimental data indicating the same mechanism, that’s when you begin to feel confident that you’ve uncovered something significant,” remarks Stokes.
For Barzilay, the initiative underscores a paradigm shift in the application of AI within the life sciences. “Much of the AI application in drug discovery has focused on exploring chemical landscapes, identifying new molecules that could be effective,” she remarks. “What we’re demonstrating here is that AI can also furnish mechanistic insights, which are essential for advancing a molecule through the developmental pipeline.”
This distinction is crucial because mechanism-of-action studies often represent a significant rate-limiting factor in drug development. Conventional approaches can extend to 18 months to two years, or more, and incur millions of dollars. In this case, the MIT–McMaster team condensed the timeline to roughly six months, at a fraction of the expense.
Enterololin is still in the nascent stages of development, but translation efforts are already in motion. Stokes’ spinout enterprise, Stoked Bio, has licensed the compound and is refining its attributes for potential human application. Preliminary research is exploring derivatives of the molecule against other resistant pathogens, such as Klebsiella pneumoniae. If all proceeds well, clinical trials could commence within the next few years.
The researchers also perceive broader ramifications. Narrow-spectrum antibiotics have long been pursued as a means to combat infections without collateral harm to the microbiome, yet they have proven difficult to discover and validate. AI tools like DiffDock could render that process more feasible, rapidly facilitating a new generation of targeted antimicrobials.
For patients with Crohn’s and other inflammatory bowel disorders, the possibility of a medication that alleviates symptoms without unsettling the microbiome could lead to a significant enhancement in quality of life. And in the broader context, precision antibiotics might contribute to addressing the escalating issue of antimicrobial resistance.
“What excites me is not solely this compound, but the concept that we can commence viewing the elucidation of mechanisms of action as something we can expedite, with the appropriate amalgamation of AI, human insight, and laboratory experimentation,” states Stokes. “That holds the potential to revolutionize our approach to drug discovery for numerous diseases, not just Crohn’s.”
“One of the most considerable challenges to our health is the rise of antimicrobial-resistant bacteria that evade even our most effective antibiotics,” adds Yves Brun, professor at the University of Montreal and distinguished professor emeritus at Indiana University Bloomington, who was not involved in the study. “AI is becoming a pivotal tool in our battle against these bacteria. This research employs a potent and sophisticated combination of AI techniques to ascertain the mechanism of action of a new antibiotic candidate, a vital stage in its prospective development as a therapy.”
Corso, Barzilay, and Stokes collaborated on the paper with McMaster scholars Denise B. Catacutan, Vian Tran, Jeremie Alexander, Yeganeh Yousefi, Megan Tu, Stewart McLellan, and Dominique Tertigas, and professors Jakob Magolan, Michael Surette, Eric Brown, and Brian Coombes. Their research received partial support from the Weston Family Foundation; the David Braley Centre for Antibiotic Discovery; the Canadian Institutes of Health Research; the Natural Sciences and Engineering Research Council of Canada; M. and M. Heersink; Canadian Institutes for Health Research; Ontario Graduate Scholarship Award; the Jameel Clinic; and the U.S. Defense Threat Reduction Agency Discovery of Medical Countermeasures Against New and Emerging Threats program.
The researchers have made the sequencing data available in public repositories and published the DiffDock-L code openly on GitHub.
“`